circos.conf
# 1.1 MINIMUM CIRCOS CONFIGURATION
#
# The ‘hello world’ Circos tutorial. Only required
# configuration elements are included.
#
# Subsequent 1.* tutorials build on this example to generate
# a representative image with common elements found in Circos
# figures in the literature.
# Chromosome name, size and color definition
karyotype = data/karyotype/karyotype.human.txt
# The <ideogram> block defines the position, size, labels and other
# properties of the segments on which data are drawn. These segments
# are usually chromosomes, but can be any integer axis.
<ideogram>
<spacing>
# Spacing between ideograms. Suffix “r” denotes a relative value. It
# is relative to circle circumference (e.g. space is 0.5% of
# circumference).
default = 0.005r
</spacing>
# Ideogram position, thickness and fill.
#
# Radial position within the image of the ideograms. This value is
# usually relative (“r” suffix).
radius = 0.90r
# Thickness of ideograms, which can be absolute (e.g. pixels, “p”
# suffix) or relative (“r” suffix). When relative, it is a fraction of
# image radius.
thickness = 20p
# Ideograms can be drawn as filled, outlined, or both. When filled,
# the color will be taken from the last field in the karyotype file,
# or set by chromosomes_colors. Color names are discussed in
#
# http://www.circos.ca/documentation/tutorials/configuration/configuration_files
#
# When stroke_thickness=0p or if the parameter is missing, the ideogram is
# has no outline and the value of stroke_color is not used.
fill = yes # A
stroke_color = dgrey # B
stroke_thickness = 2p # B
</ideogram>
################################################################
# The remaining content is standard and required. It is imported from
# default files in the Circos distribution.
#
# These should be present in every Circos configuration file and
# overridden as required. To see the content of these files,
# look in etc/ in the Circos distribution.
<image>
# Included from Circos distribution.
<<include etc/image.conf>>
</image>
# RGB/HSV color definitions, color lists, location of fonts, fill patterns.
# Included from Circos distribution.
<<include etc/colors_fonts_patterns.conf>>
# Debugging, I/O an dother system parameters
# Included from Circos distribution.
<<include etc/housekeeping.conf>>
shenzy@shenzy-ubuntu:/winxp_disk2/shenzy/circos/circos-tutorials-0.62/tutorials/1/1$ ../../../../circos-0.62-1/bin/circos -conf ./circos.conf
debuggroup conf 0.13s welcome to circos v0.62-1 25 Jun 2012
debuggroup conf 0.13s loading configuration from file ./circos.conf
debuggroup conf 0.13s looking for conf file ./circos.conf
debuggroup conf 0.13s found conf file ./circos.conf
debuggroup summary 0.31s debug will appear for these features: summary
debuggroup summary 0.31s parsing karyotype and organizing ideograms
debuggroup summary 0.49s applying global and local scaling
debuggroup summary 0.51s allocating image, colors and brushes
debuggroup summary 1.43s drawing highlights and ideograms
debuggroup summary,output 1.56s generating output
debuggroup summary,output 2.13s created PNG image ./circos.png (56 kb)
debuggroup summary,output 2.13s created SVG image ./circos.svg (11 kb)
shenzy@shenzy-ubuntu:/winxp_disk2/shenzy/circos/circos-tutorials-0.62/tutorials/1/2$ cat circos.conf
# 1.2 IDEOGRAM LABELS, TICKS, AND MODULARIZING CONFIGURATION
#
# In this tutorial, I will add tick marks, tick labels and ideogram
# labels to the previous image. This will require the use of a <ticks>
# block and expanding the <ideogram> block.
#
# To make the configuration more modular, the tick and ideogram
# parameters will be stored in different files and imported using the
# <<include>> directive.
#
karyotype = data/karyotype/karyotype.human.txt
# The chromosomes_unit value is used as a unit (suffix “u”) to shorten
# values in other parts of the configuration file. Some parameters,
# such as ideogram and tick spacing, accept “u” suffixes, so instead of
#
# spacing = 10000000
#
# you can write
#
# spacing = 10u
#
# See ticks.conf for examples.
chromosomes_units = 1000000
<<include ideogram.conf>>
<<include ticks.conf>>
<image>
<<include etc/image.conf>>
</image>
<<include etc/colors_fonts_patterns.conf>>
<<include etc/housekeeping.conf>>
shenzy@shenzy-ubuntu:/winxp_disk2/shenzy/circos/circos-tutorials-0.62/tutorials/1/2$ cat ideogram.conf
<ideogram>
<spacing>
default = 0.005r
</spacing>
# Ideogram position, fill and outline
radius = 0.90r
thickness = 20p
fill = yes
stroke_color = dgrey
stroke_thickness = 2p
# Minimum definition for ideogram labels.
show_label = yes
# see etc/fonts.conf for list of font names
label_font = default
label_radius = dims(image,radius) – 60p
label_size = 30
label_parallel = yes
</ideogram>
shenzy@shenzy-ubuntu:/winxp_disk2/shenzy/circos/circos-tutorials-0.62/tutorials/1/8$ ll
total 3431
-rwxrwxrwx 1 root root 315 2012-06-22 07:33 axes.conf
-rwxrwxrwx 1 root root 331 2012-06-22 07:34 backgrounds.conf
-rwxrwxrwx 1 root root 3584 2012-09-11 15:24 circos.conf
-rwxrwxrwx 1 root root 1302846 2012-09-11 15:25 circos.png
-rwxrwxrwx 1 root root 2188271 2012-09-11 15:25 circos.svg
-rwxrwxrwx 1 root root 51 2012-09-11 15:25 exclude.hs1.rule
-rwxrwxrwx 1 root root 450 2012-06-26 04:49 ideogram.conf
-rwxrwxrwx 1 root root 709 2012-06-09 08:22 ticks.conf
shenzy@shenzy-ubuntu:/winxp_disk2/shenzy/circos/circos-tutorials-0.62/tutorials/1/8$ more circos.conf
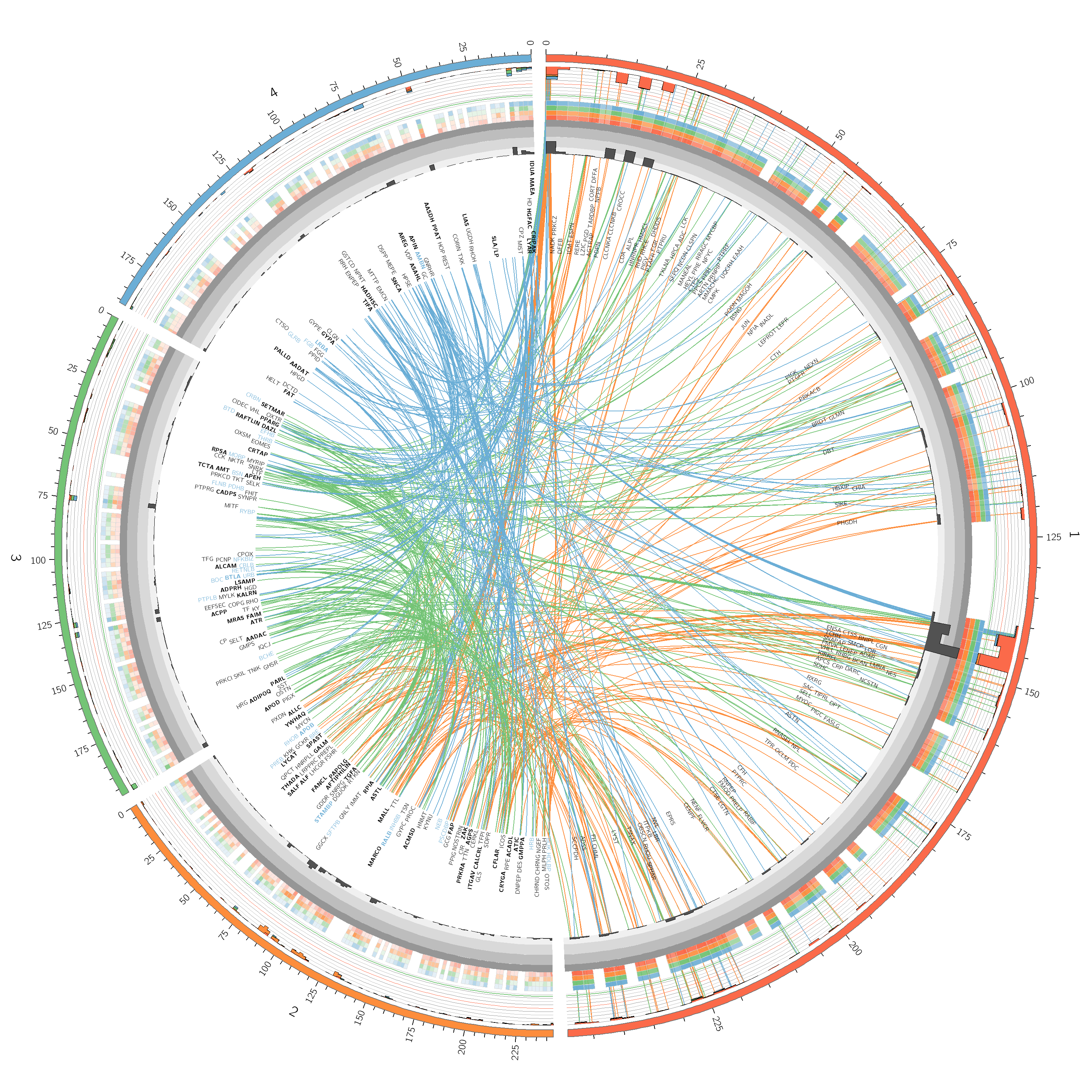
# 1.8 TEXT TRACKS
karyotype = data/karyotype/karyotype.human.txt
chromosomes_units = 1000000
chromosomes_display_default = no
chromosomes = /hs[1234]$/
<colors>
chr1* = red
chr2* = orange
chr3* = green
chr4* = blue
</colors>
chromosomes_reverse = /hs[234]/
chromosomes_scale = hs1=0.5r,/hs[234]/=0.5rn
<plots>
<plot>
type = text
file = data/6/genes.labels.txt
# Like with other tracks, text is limited to a radial range by setting
# r0 and r1.
#
# Individual labels can be repositioned automatically with in a
# position window to fit more labels, without overlap. This is an
# advanced feature – see the 2D Track text tutorials.
r1 = 0.8r
r0 = 0.6r
# For a list of fonts, see etc/fonts.conf in the Circos distribution.
label_font = light
label_size = 12p
# padding – text margin in angular direction
# rpadding – text margin in radial direction
rpadding = 5p
# Short lines can be placed before the label to connect them to the
# label’s position. This is most useful when the labels are
# rearranged.
show_links = no
link_dims = 0p,2p,5p,2p,2p
link_thickness = 2p
link_color = black
<rules>
<<include exclude.hs1.rule>>
# Text can be tested with var(value).
<rule>
condition = var(value) =~ /a/i
label_font = bold
flow = continue
</rule>
<rule>
condition = var(value) =~ /b/i
color = blue
</rule>
</rules>
</plot>
<plot>
type = heatmap
file = data/5/segdup.hs1234.heatmap.txt
r1 = 0.89r
r0 = 0.88r
color = hs1_a5,hs1_a4,hs1_a3,hs1_a2,hs1_a1,hs1
scale_log_base = 5
<rules>
<<include exclude.hs1.rule>>
<rule>
condition = var(id) ne “hs1″
show = no
</rule>
</rules>
</plot>
<plot>
type = heatmap
file = data/5/segdup.hs1234.heatmap.txt
r1 = 0.90r
r0 = 0.89r
color = hs2_a5,hs2_a4,hs2_a3,hs2_a2,hs2_a1,hs2
scale_log_base = 5
<rules>
<<include exclude.hs1.rule>>
<rule>
condition = var(id) ne “hs2″
show = no
</rule>
</rules>
</plot>
<plot>
type = heatmap
file = data/5/segdup.hs1234.heatmap.txt
r1 = 0.91r
r0 = 0.90r
color = hs3_a5,hs3_a4,hs3_a3,hs3_a2,hs3_a1,hs3
scale_log_base = 5
<rules>
<<include exclude.hs1.rule>>
<rule>
condition = var(id) ne “hs3″
show = no
</rule>
</rules>
</plot>
<plot>
type = heatmap
file = data/5/segdup.hs1234.heatmap.txt
r1 = 0.92r
r0 = 0.91r
color = hs4_a5,hs4_a4,hs4_a3,hs4_a2,hs4_a1,hs4
scale_log_base = 5
<rules>
<<include exclude.hs1.rule>>
<rule>
condition = var(id) ne “hs4″
show = no
</rule>
</rules>
</plot>
<plot>
type = histogram
file = data/5/segdup.hs1234.hist.txt
r1 = 0.88r
r0 = 0.81r
fill_color = vdgrey
extend_bin = no
<rules>
<<include exclude.hs1.rule>>
</rules>
<<include backgrounds.conf>>
</plot>
<plot>
type = histogram
file = data/5/segdup.hs1234.stacked.txt
r1 = 0.99r
r0 = 0.92r
fill_color = hs1,hs2,hs3,hs4
orientation = in
extend_bin = no
<rules>
<<include exclude.hs1.rule>>
</rules>
<<include axes.conf>>
</plot>
</plots>
<links>
<link>
file = data/5/segdup.txt
radius = 0.6r
bezier_radius = 0r
color = black_a4
thickness = 2
<rules>
<rule>
condition = var(intrachr)
show = no
</rule>
<rule>
condition = 1
color = eval(var(chr2))
flow = continue
</rule>
<rule>
condition = from(hs1)
radius1 = 0.99r
</rule>
<rule>
condition = to(hs1)
radius2 = 0.99r
</rule>
</rules>
</link>
</links>
<<include ideogram.conf>>
<<include ticks.conf>>
<image>
<<include etc/image.conf>>
</image>
<<include etc/colors_fonts_patterns.conf>>


Recent Comments